experimental mapping of all Arabidopsis transcription units using Affymetrix genome tiling arrays
Overview of the Project
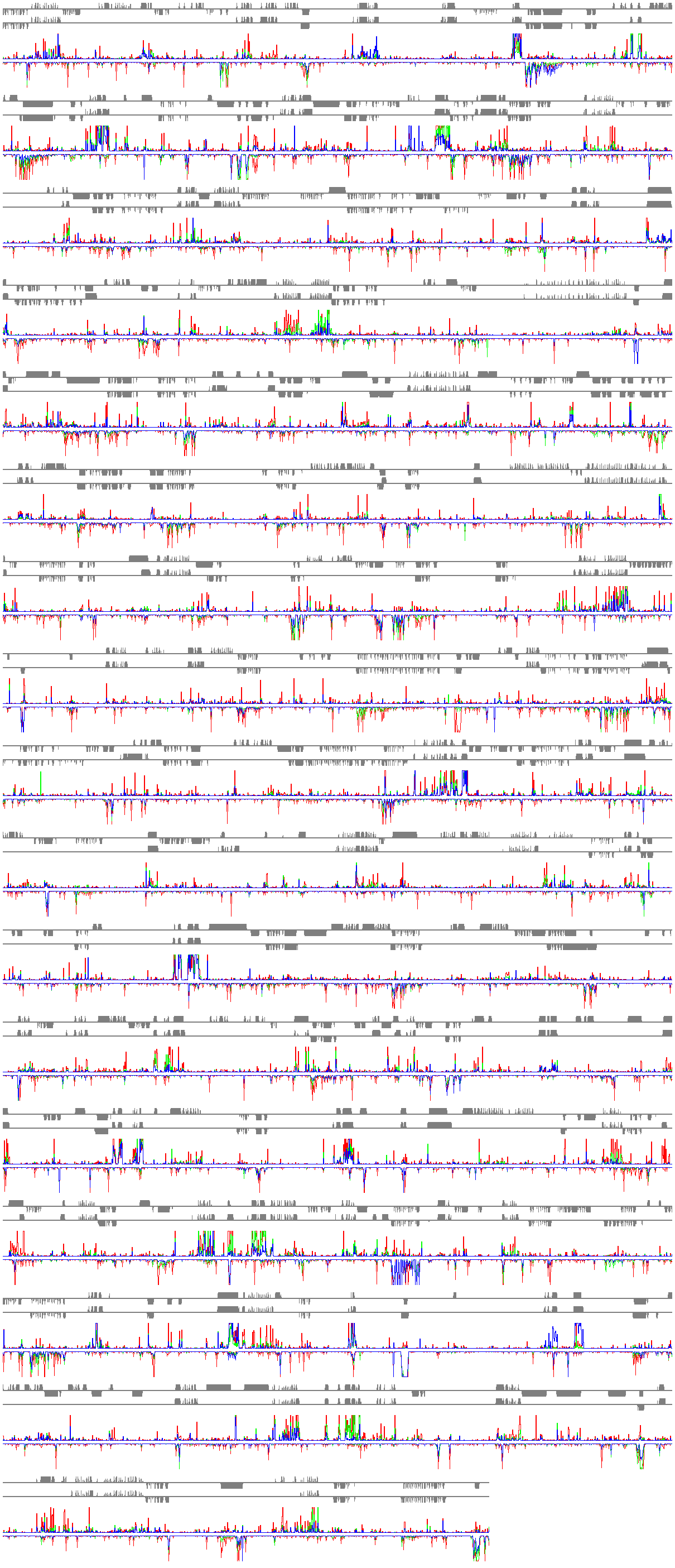
Functional genomic analysis of any organism with a complete genome sequence will require accurate gene structure information and a complete gene inventory. Using the reference plant Arabidopsis, collections of full-length (fl) cDNAs are being sequenced by the Salk (J. Ecker), Stanford (R. Davis), Plant Gene Expression Center/Berkeley (A. Theologis) Consortium. The fl-cDNA sequence information is being used to correct our initial computational gene predictions in the genome sequence (1). However, even after extensive cDNA library development and ESTs sequencing, fl-cDNAs have been discovered for only about two-thirds of the ~25,500 predicted Arabidopsis genes (2). Therefore, novel approaches are required to verify the accuracy of genome annotation and more importantly, to discover new (unpredicted) genes in genome sequence. Utilizing a suite of custom high-density (~832,00 25-mer oligonucleotides/chip) Affymetrix arrays and in-house Chipviewer visualization/analysis software we are carrying out empirical annotation of the transcriptional activity in the entire Arabidopsis genome sequence. In brief, labeled cRNA preparations from various tissues, and treatments is hybridized to a variety of different types of custom high-density genome tiling arrays, include a complete end-to-end genome tile. This unbiased approach allows accurate determination of the gene structures, and identification of thousands of novel transcription units. The chip data are being used as a guide to isolate fl-cDNAs for each of these "untouched" Arabidopsis genes.
1. The Arabidopsis Genome Initiative, Nature 408, 796 (2000)
2. M. Seki et al., Science 296, 141 (2002).
1. The software.
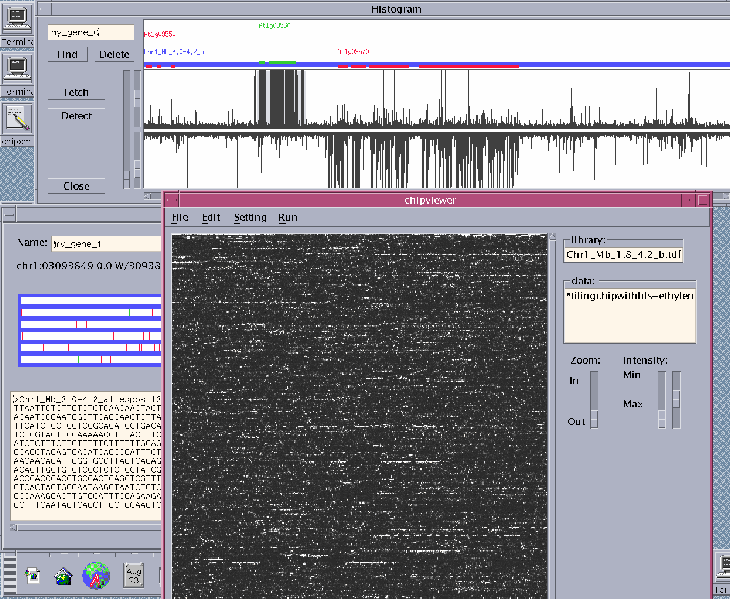
2. The signals